Proteinase-Activated Receptor 2 Antibodies
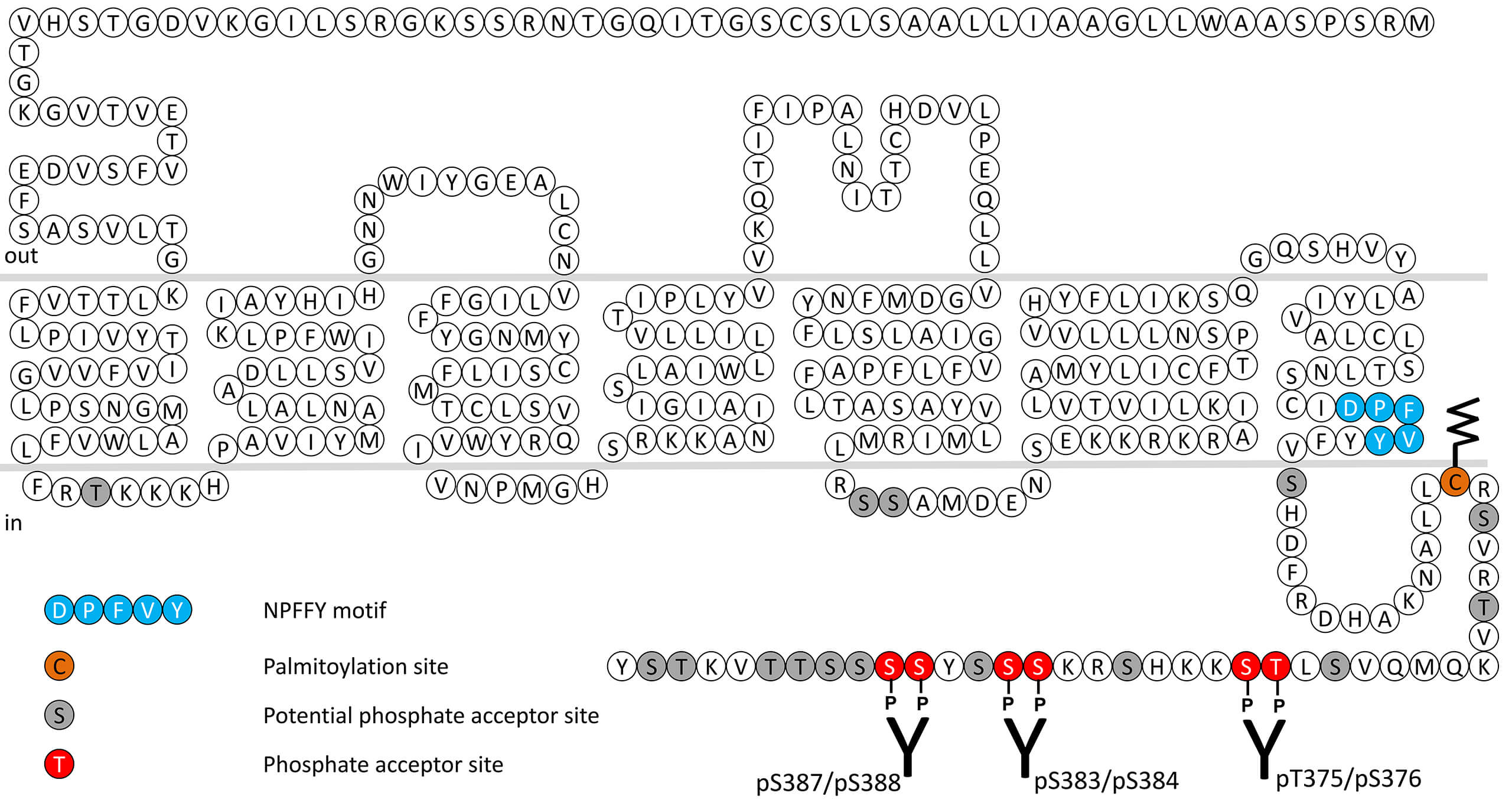
Proteinase-activated receptors (PARs) are unique members of the GPCR superfamily activated by proteolytic cleavage of their amino terminal exodomains. Agonist proteinase-induced hydrolysis unmasks a tethered ligand (TL) at the exposed amino terminus, which acts intramolecularly at the binding site in the body of the receptor to effect transmembrane signaling. TL sequences at human PAR1-4 are SFLLRN-NH2, SLIGKV-NH2, TFRGAP-NH2 and GYPGQV-NH2, respectively. Several proteinases, including neutrophil elastase, cathepsin G and chymotrypsin can have inhibitory effects at PAR1 and PAR2 such that they cleave the exodomain of the receptor without inducing activation of Gαq-coupled calcium signaling, thereby preventing activation by activating proteinases but not by agonist peptides. Neutrophil elastase (NE) cleavage of PAR1 and PAR2 can however activate MAP kinase signaling by exposing a TL that is different from the one revealed by trypsin. PAR2 activation by NE regulates inflammation and pain responses and triggers mucin secretion from airway epithelial cells. PAR2 receptor desensitization and internalization are regulated by phosphorylation of carboxyl-terminal threonine375/serine376 (pT375/pS376-PAR2), serine382/serine384 (pS383/pS384-PAR2) and serine387/serine388 (pS387/pS388-PAR2). This nomenclature refers to the human PAR2 receptor. This phosphorylation motif is conserved across species and is identical in rats and corresponds to pS377/pS378-PAR2 and pS389/pS390-PAR2 in mice. For more information on PAR2 pharmacology please refer to the IUPHAR database. For further reading refer to:
Hollenberg MD, Compton SJ. International Union of Pharmacology. XXVIII. Proteinase-activated receptors. Pharmacol Rev. 2002 Jun;54(2):203-17. doh: 10.1124/pr.54.2.203. PMID: 12037136.
Lin H, Liu AP, Smith TH, Trejo J. Cofactoring and dimerization of proteinase-activated receptors. Pharmacol Rev. 2013 Sep 24;65(4):1198-213. doi:10.1124/pr.111.004747. PMID: 24064459; PMCID: PMC3799237.
Bunnett N, DeFea K, Hamilton J, Hollenberg MD, Ramachandran R, Trejo J. Proteinase-activated receptors (version 2019.4) in the IUPHAR/BPS Guide to Pharmacology Database. IUPHAR/BPS Guide to Pharmacology CITE. 2019; 2019(4). Available from: https://doi.org/10.2218/gtopdb/F59/2019.4.
 pT375/pS376-PAR2 (phospho-Proteinase-Activated...
pT375/pS376-PAR2 (phospho-Proteinase-Activated...  pS383/pS384-PAR2 (phospho-Proteinase-Activated...
pS383/pS384-PAR2 (phospho-Proteinase-Activated...  pS387/pS388-PAR2 (phospho-Proteinase-Activated...
pS387/pS388-PAR2 (phospho-Proteinase-Activated...